How DNAcrypt-AI works
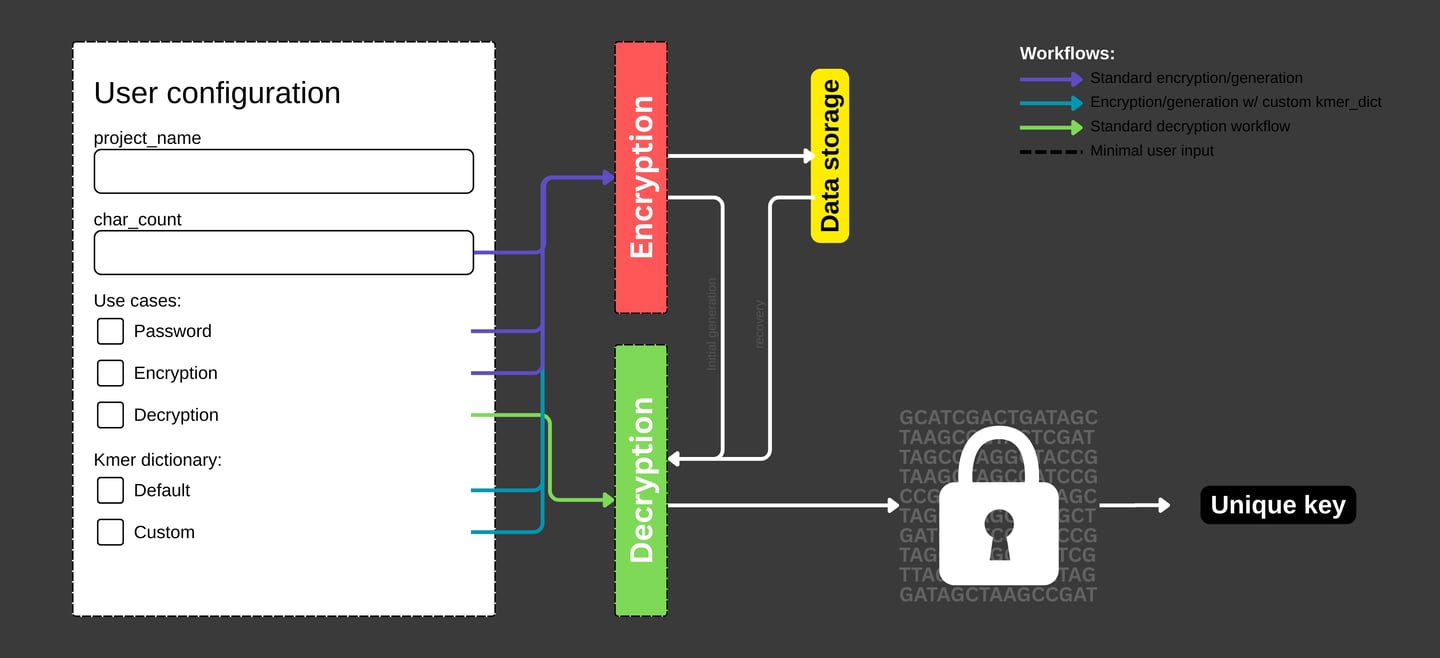
Quick Overview
DNAcrypt-AI generates a random password (alphanumeric + symbols) or cryptographic key (alphanumeric only) based on a user-defined length. The generated password or key is encrypted by mapping it to coordinates of randomly sampled, variable-length DNA sequences from the human genome, referenced to the hg19 and hg38 assemblies.
To decrypt the information, DNAcrypt-AI reconstructs the corresponding DNA sequences using a high-throughput sequence reconstitution pipeline (FAS2rDNA) and interprets them with a sequence-informed machine learning model (Covary).
Supported Encodings
DNAcrypt-AI supports the following character encodings:
Alphanumeric (Passwords & Keys):
a–z, A–Z, 0–9Symbols (Passwords only):
! @ # $ % ^ & * ( ) - _ + = ”
Human Genome Assemblies
DNAcrypt-AI uses the hg19 and hg38 human genome assemblies as reference spaces for encryption and decryption. These assemblies provide the biological sequence coordinates used to store and retrieve encrypted information. Work is currently underway to expand support to multi-species genome assemblies, which will further increase the genome vocabulary and entropy of DNAcrypt-AI. Suggestions and contributions to improve this capability are welcome.
Encrypting a password or key
DNAcrypt-AI is designed to be intuitive and easy to use, as a Jupyter notebook in Google Colab. To generate and encrypt a password or key, users follows the procedure below:
Create a user configuration
Set char_count to define the desired length
Select a Use case:
Password: alphanumeric + symbols
Encryption: alphanumeric only
Run DNAcrypt-AI
Select Runtime → Run all
Download and store your encrypted data
The following files will be generated:
DNAcrypt_metadata.json
kmer_dict.json (only if a custom k-mer dictionary is used)
The enrypted files are automatically downloaded. If your browser blocks downloads, you can retrieve them through the File browser manually from:
/content/
/content/DNAcrypt/outputs/
Decrypting a password or key
Decrypting your data is straightforward:
Modify the user configuration
Set the Use case to Decryption
Run DNAcrypt-AI
Select Runtime → Run all
Upload your encrypted data
Always upload DNAcrypt_metadata.json
Upload kmer_dict.json only if a custom k-mer dictionary was used during encryption
Wait for decryption to finish
Decryption typically completes within 15 minutes or less, depending on the size of the genome vocabulary used
Using a custom kmer_dict
Custom kmer_dict (kmer dictionary) allows users to vary the kmer-to-charcter encodings, providing them with the ability to customized their character dictionaries. This feature is valuable for users to serve as second layer of encryption, refactor their compromised data to generate new sequence, or customize their library for specific needs.
A. During encryption
In the user configuration:
Set char_count
Select the appropriate use case (Password or Encryption)
Choose Custom under K-mer Dictionary
Run DNAcrypt-AI as usual
Store the following files for future recovery:
DNAcrypt_metadata.json
kmer_dict.json
B. During decryption
Select Decryption as the use case
Run DNAcrypt-AI
Upload both:
DNAcrypt_metadata.json
kmer_dict.json
Handling and storing encrypted data
Your encrypted files must not be modified. Any loss, alteration, or unintended addition may prevent successful decryption. Tampering with either DNAcrypt_metadata.json or kmer_dict.json will affect recovery of your password or key.
File names may be changed, but file contents must remain intact.
Encrypted data is lightweight (typically under 200 KB) and can be stored in several ways:
Printed copy: Highly private and offline, but requires re-encoding into digital form before use
Digital copy: Immediately usable, but anyone with access to the files can attempt decryption