DNAcrypt-AI architecture
Overview
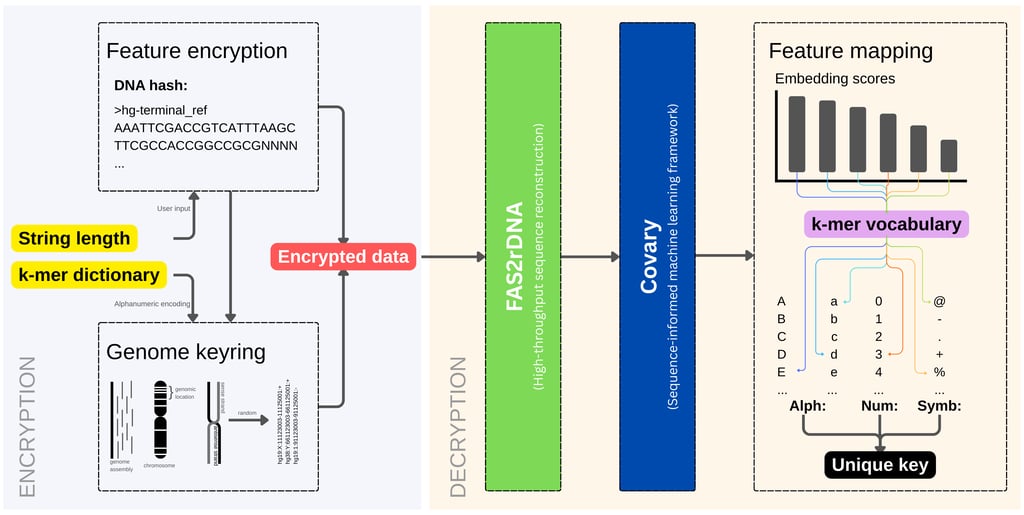
DNAcrypt-AI is a human genome–based cryptographic system built on two genome intelligence frameworks, FAS2rDNA and Covary. At its core, DNAcrypt-AI exploits the intrinsic complexity, scale, variation, and unpredictability of the human genome to generate, encrypt, and decrypt passwords and cryptographic keys in a biologically grounded and AI-driven manner.
Design Principles
DNAcrypt-AI operates by constructing a genome vocabulary randomly sampled from more than three billion (3B) genomic loci across multiple chromosomes and human genome assemblies (hg19 and hg38). This vocabulary consists of genomic coordinates, forming the encrypted representation of the secret data.
These coordinates are interpreted by FAS2rDNA, which reconstructs DNA sequences typically ranging from 1,000 to 50,000 nucleotides in length. The resulting sequences form a heterogeneous genomic corpus, aggregated into 1,025 vocabularies. The resulting genome sequence vocabulary is massively large, structurally diverse, and resistant to direct interpretation, visual inspection, or conventional bioinformatics analysis.
Sequence Intelligence
Once reconstructed, the genomic vocabulary is processed by Covary, a sequence-informed machine learning framework designed to extract latent relational patterns from biological sequences.
Covary encodes the reconstructed DNA using feature expansion derived from TIPs-VF (refactored as Covary_encoder), enabling pattern extraction without reliance on frequency-based or reducible signatures. This design ensures that the transformation pipeline remains traceable but non-reversible, a key requirement for cryptographic robustness. The output of Covary is a set of vector embedding scores that represent deep, learned sequence relationships that can be used to map the embeddings with the alphanumeric characters and symbols that are encoded in a kmer dictionary.
Internal workflow & process
1. User input: The user specifies a workflow and desired key length.
Alphanumeric only (encryption key)
Alphanumeric + symbols (password)
Supported lengths range from 6 to 90 characters, with a default of 12.
2. Encoding and k-mer mapping: A predefined k-mer dictionary (k = 5) encodes characters into a non-reversible mapping space.
Users may optionally generate custom k-mer dictionaries with dynamic character-to-k-mer mappings to increase variability and security.
3. Genome keyring: Deep random sampling of genomic coordinates from different human genome assemblies, chromosome identifiers, strand orientation, and start-end genomic locations.
4. Genome vocabulary expansion: DNAcrypt-AI aggregates genome-derived vocabularies into a corpus containing up to 51 million (51 M) nucleotides.
5. Sequence reconstruction and learning
FAS2rDNA reconstructs DNA sequences from genomic coordinates using a high-throughput, multiprocessing pipeline.
Covary analyzes the reconstructed corpus and learns relational sequence patterns.
6. Cryptographic transformation: DNAcrypt-AI consumes the vector embeddings produced by Covary to map embedding scores and/or angular derivatives back onto the encodings in the k-mer dictionary.
This final transformation yields a unique cryptographic sequence, which is deterministically decrypted using the DNA hash to recover the original password or encryption key.